
Alive and Inactivated Cutibacterium Acnes: Properties, Functions and Pathogenicity
*Corresponding Author(s):
Maria VadalàMedico Cura Te Stesso Onlus, Modena (MO), Via Ciro Bisi 125, Italy
Email:mary.vadala@gmail.com
Abstract
Cutibacterium acnes (C.acnes) is a lipophilic member of the human skin microbiota, usually colonizing the infundibular spaces of the sebaceous glands, lipids particularly rich.
We have tried to summarize in this paper the main biological traits of the alive C.parvum strain in a clinical perspective, in order to better clarify and take advantage of the complex interaction between its functional subcellular components and the host cells either into the skin or in the deep organs to achieve or maintain the best healthy conditions.
Introduction
Cutibacterium acnes (C.acnes) is a Gram-positive lipophilic member of the human skin microbiota into the lipids filled sebaceous, glands, almost colonizing the infrainfundibular spaces.
More recently, it has also been found in several other organs and tissue sites on epithelial or mucosal linings, such as oral cavity, stomach, lung, urinary tract, and prostate [1-4].
It is thus now debatable whether it is a resident of healthy human skin, or if it can spontaneously saprophytize multiples organs, taking advantage of temporary skin disruption or failure of the host immunosurveillance.
As a matter of fact, C.parvum can be a facultative pathogen in the following diseases:
Progressive macular hypolmelanosis (PMH), smooth hypopigmented skin areas rich in sebaceous areas; often the lower back skin is affected, the strains involved belong to phylotype III (SLST type L) of C. acnes (renamed to C. acnes subsp. Elongatum) [5,6].
Acne fulminans, with heavy ulcerated comedones and spreading infectious symptoms recognizes, but with low evidence the SLST type A (phylotype IA1) in about 60% of patients [7].
Prosthetic infections: more and more studies report the increased detection of C.acnes in prosthetic implants in the last years [8-10].
Sonication of periprosthetic granulation tissue to be cultured allowed probably to identify the bacterium with the aid of proper culture techniques acnes, (e.g. cultivation times have often been extended to 14 and even 21 days) [11,12]. Also, the diagnostic mass spectrometry “Matrix Assisted Laser Desorption Ionization-Time of Flight” (MALDI-TOF) as well as the Polymerase chain reaction (PCR) techniques are very much helpful to simplify the identifications o low aggressive strains.
As to the prosthetic joint infections, the 10% find C.parvum into the environment, particularly in the shoulder (31-70% of all joint infections) which being contiguous to axillary region rich in sebaceous glands, the men are more exposed to C.acnes due to the different anatomic distribution of pilosebaceous units [13,14]. Also spinal instrumentation surgeries bring some more risk of C.acnes contamination [15]. The symptoms can be quite weak (mild pain, no fever) and mainly local lasting 2 or more years; mobility of the prosthesis and radiographic signs (osteolytic shadows) are quite late. Prosthetic heart valves and rings, or implantable pacemakers can cause C.acnes-associated infective endocarditis (IE) with infectious symptoms and uneasy culture diagnosis.
In breast prosthesis C.acnes colonization and biofilm is associated with capsular contracture [16].
Lee et al. [17] confirmed that C.acnes was found as the most prevalent microorganism in cases with chronic infection.
In 15% of infections associated with implanted drain cerebrospinal fluid (CSF) tubes to the peritoneum with low fever and inadvertent clinical symptoms [18,19], PCR on CSF (a good, even too much sensitive detection tool) demonstrated C.parvum colonization [20].
Generally speaking, only high repeated titres of positive C.acnes CFU in a certain number of samples from the infection site state it’s causative role, while a single positive culture should be taken with caution.
Native or acquired spine infections, spine surgery, and spine disk degeneration (especially Modic type 1 disc atrophy characterized by fissuring and edema of the endplates) often suggest C.acnes as a putative cause, being such infections successfully treated with antibiotic therapy [21].
Prostate Pathologies
C. acnes has been found in prostate inflammation and prostate cancer [22]. A study from Sweden reported that C. acnes was cultured in 60% of the prostate cancer cases (n=100) and in 26% of cancer-free controls (n=50) [4]. The phylotype II (SLST type K) was the most dominant type among C. acnes strains obtained from prostatic tissue and 26% of those strains carried an extra chromosomal element [23]. It is debated if they represent contamination/carry-over from the urogenital tract, or if they are colonizers of the tumour tissue (anoxic regions). In contrast a French study has detected only very few C.acnes positive samples in their cohort (n=36) [24].
Sarcoidosis
Sarcoidosis is non-necrotizing granulomatous on a systemic background located mainly in the lungs, but also in the skin, lymph nodes, eyes, and other body locations. The most frequent infectious agents of sarcoidosis are primarily Mycobacterium tuberculosis and C.acnes [25,26].
C.acnes experimentally reproduced sarcoidosis in mice [27,28], hypothetically C.acnes trespassed the disrupted skin barrier could colonize intracellularly macrophages like Trojan horses; insoluble immune complexes are then generated, due to a possible genetic individual Th1 cells hypersensitivity against C. acnes [26].
Several recent studies have identified C.acnes without specific C.acnes phylotype in human sarcoidotic granulomas by means of PCR, immunohistochemistry/immunofluorescence staining [29-31].
Epithelioid palisades and giant cells concur to generate the pathologic background of sarcoid nodules which usually remit with different protocols of steroid therapy.
C.acnes Infection and Virulence Factors: Alive Versus Killed C.parvum
The virulence factors of C.acnes, that will be later presented in details, play a role in the pathogenesis of human diseases, that can be actively or passively secreted, integrated in the cell membrane either attached or embedded: they are Christie–Atkins–Munch-Petersen (CAMP) factors, dermatan-sulfate adhesins (DsAs), lipases, sialidases, hyaluronidases, putative endoglycoceramidases, porphyrins, short-chain fatty acids (SCFAs), cell wall polysaccharide/lipoglycan, and lipoproteins [22,32-38]. We have no evidence from our experience and literature review that subcutaneous injection of the killed C.acnes might be involved or promote any bacterium-dependent pathology; on the contrary accordingly with the mice protection experimental study [39], the killed Corynebacterium injection, or C.parvum sialidase based vaccine is protective against C.acnes challenges and it reduces also its inflammatory cytokines burst.
In fact, it releases a complex cascade of subcellular components challenging the innate immunity signals and promotes killer lymphocytes and phagocyting cells (monocytes, macrophages dendritic and apc cells) functional enhancement through inflammatory cytokines and interferons release.
This curative approach has effectively been addressed in the past to fight cancer, viral and other pathogenic agents infections and, respectively, it might also prevent infections of the same alive bacterium as successfully previously attempted with C.parvum based anti-acnes vaccines.
Morphological Details of C.parvum (C.acnes)
Ultrastructurally, it has an unique cell wall made of specific D-alanyne and L-acid diaminopleic rich peptidoglycans and envelope, containing phosphatidylinositol, triacylglycerol, and other common lipids [40].
Analyses of C.acnes lipoglycans have also revealed the presence of a lipid anchor based on fatty acids and shown that the polysaccharide moiety contains significant amounts of mannose, glucose, and galactose, and diaminohexuronic acid [41].
Morphologically, its appearance is somehow rod-shaped and slightly curved measuring 0.4 to 0.7µm x 3 to 5µm, mimicking diphtheroid or coryneform bacteria. It is classified as “aerotolerant anaerobe” because of oxygen detoxifying enzymes [42], oxidative phosphorylation (NAPDH dehydrogenase/complex I, cytochrome d oxidase genes, cytochrome c reductase, cytochrome c oxidase, and FoF1-type ATP synthase), this explains its growth in anaerobic conditions and a much slower growing capacity in aerobiosis [43].
Genetic Details
Genus Cutibacterium belongs to a branch of actinobacter that beyond the sin species subgroup encloses also “Propionibacterium freundenreichii” a Swiss cheese conditioner producing propionic acids, very much focused in the zootechnical setting because of its bovine rumen functions improving.
The next generation sequencing (NGS) has added a genetic detail about C.acnes and the skin microbiome, whose genome was sequenced in its entirety in 2004. It is a single circular chromosome of 2,560,265 base pairs corresponding to 2333 potential genes [32].
During the past decade, several biochemical, transcriptomic, and proteomics analyses have shown that the various phylotypes of C.acnes have different inflammatory potentials and express different putative virulence factors.
Summarizing, it has a core genome phylogeny, with 10 lineages (types A-L); the complex phylotype IA1 is further split into five SLST types (A-E) [44]. The other SLST types phylotypes are subdivided as follows: F, IA2; G, IC; H, IB; K, II; L, III (Figure 1).
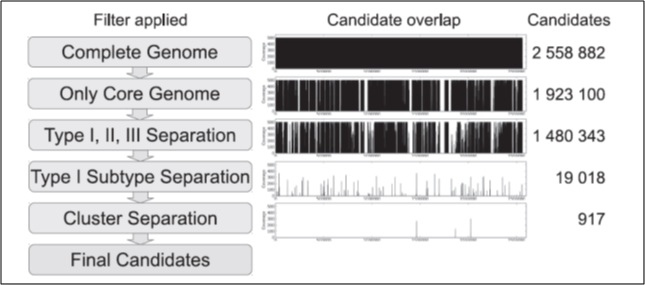
Figure 1: Strategy for the identification of SLST candidates in P.acnes. The first column shows the applied filter. The second column indicates the location of the remaining candidate fragments across the genome. The third column gives the number of remaining candidates. Copyright 2014. Published by Plos One, All rights reserved.
The accessory genome of C.acnes is not large and encloses a linear and a circular plasmid expressing resistance to macrolides, clindamycin and tetracycline [23,45,46]. 60 other regions outside of the core genome can be identified in the pan-genome of C.acnes code for multiples enzymatic functions [45,47,48], leading to bacteriocins synthesis, resistance to heavy metals and other chemical compounds etc.
C.acnes has been considered a commensal bacterium for a long time, but its implication in various types of infection qualified it as an opportunistic, low pathogenic, pathogen. Several molecular mechanisms enclosed the expression of virulence factors are involved in the adaptation process over the skin or more deeply into the host with facultative damage and triggering innate immune response as well.
C.acnes can release lytic enzymes (metalloproteases, lipases, proteases, hyaluronidases) into its environment, and these enzymes can provide disruption of the follicular epithelium and activation of the immune system [48].
Various putative virulence factor genes have been identified in the C.acnes genome. Some may be involved in cell adhesion, whereas others may mediate inflammation, tissue invasion/degradation in the host, and the synthesis of capsule polysaccharides; they are: Lipases sialidases, neuraminidases, endoglycoceramidases, adhesins, thermal shock proteins, CAMP factor, lipases/esterases and porphyrin [32,49].
LIPASES: The genome of C.acnes encodes at least 12 putative lipases probably located on the surface of the bacterial membrane [50]. A triacylglycerol lipase, glycerol-ester hydrolase A (GehA), in particular was the first molecule identified as putative C.acnes virulence factors, because it produces free fatty acids (FFAs), thereby promoting inflammation.
PUFA: The polyunsaturated fatty acid (PUFA) isomerase from C.acnes is a yellow 424-residue monomeric protein that can catalyse the isomerization of conjugated linoleic acid (CLA). CLA and its isomers regulate several functions in humans and are present in low levels in food [51].
Hyaluronate lyase (HYL) degrades hyaluronic acid (HA) and other glycosaminoglycans (GAG), such as chondroitin-4-sulfate, chondroitin-6-sulfate, and dermatan sulfate, present in the extracellular matrix of the epidermis and dermis. It is a virulence factor promoting the spread of inflammation through the extracellular matrix (ECM) depolymerization which facilitates the spread of infectious agents. HA degradation metabolites by HYL are also medium growth factors and proinflammatory compounds as well.
Glycosidase enzymes break down glycolipids and glycoproteins and are subdivided in 2 groups: 1) Exo- and endoglycosidases, responsible for hydrolysing neutral sugars; 2) Sialidases, break down electron negative the neuraminic acid or sialic acid: Sialidases hydrolyses sialic acid from sialo glycoconjugates.
The C.acnes genome contains three genes encoding highly immunogenic sialidases/neuraminidases that increase pathogenicity of the C.parvum strains.
Sortases are covalent linking proteolytic enzymes degrading adhesion factors, to the cell walls of Gram-positive bacteria.
Porphyrins are fluorescent molecules produced by both eukaryotic and prokaryotic cells. Coproporphyrin III is present in large amounts in acne lesions [52], and type I C.acnes strains produce significantly more porphyrins than other phylotypes.
Biofilms are an extracellular matrix melting polysaccharides, proteins, and/or extracellular DNA produced endogenous and exogenously by the bacteria [53].
The C.acnes biofilm contains mostly poly-β (1-6)-N-acetylglucosamine (PNAG) polysaccharides, proteins, including the GroEL chaperonin, the elongation factors EF-Tu and EF-G, and many enzymes.
Studies of a polymicrobial biofilm mixing C.acnes and Candida albicans have shown that the latter has a protective effect on the former, reducing by 40% the lethality of C.acnes strains [54].
Sometimes, C.acnes colonizes human bone marrow-derived mesenchymal stem cells switching from commensal lifestyle to opportunistic pathogen, by increasing biofilm formation [55].
RoxP, the radical oxygenase of C.acnes, reduces free radicals enzymatically. This ROS-scavenging enzyme was purified from C.acnes for the first time in 2016. It is present only in C.acnes and is secreted into the supernatant culture. RoxP has recently been shown to protect skin cells against oxidative stress [56], RoxP is currently thought to help C.acnes to survive in oxygen-rich environments, such as the skin surface [57].
DsA1 Nine putative adhesion protein genes have been identified in the C.acnes that codify the molecular microbic surface components recognize adhesive molecules of the matrix (MSCRAMM). These adhesion proteins are truly pathogenic take part in biofilm formation, genome. One of these genes encodes the DsA1 protein, both secreted and cell-wall anchored, which binds dermatan sulphate [58] and has also been characterized as a fibrinogen-binding protein. It appears to be highly glycosylated and contains an N-acetylgalacosamine (GlaNAc) residue.
Fibrinogen adheres to the surface of C.acnes and mediates platelet aggregation [59]. DsA1 therefore appears to be an important surface protein expressed by C.acnes and further investigations of its role as a virulence factor are required.
CAMP Factors: are toxin proteins secrete or attached to the cell surface that form pores in host membranes, leading to host tissue damage and stimulating the innate immune system through CAMP factors. CAMP1 and CAMP2 are the predominant CAMP factors produced by C.acnes strains, the CAMP1-TLR2 interaction is different between phylotypes IA, II, and III (Figure 2). CAMP1 also displays a high degree of genetic polymorphism, with 14 strain-specific amino acid changes inducing strong inflammation and the production of a CAMP1 factor strongly recognized by TLR2 [60].
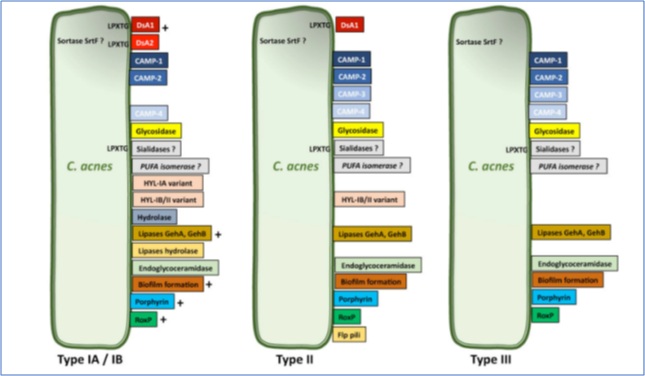 Figure 2: Differential expressions of virulence factor by C.acnes. Copyright 2021 MDPI. Published by Microorganisms, All rights reserved.
Figure 2: Differential expressions of virulence factor by C.acnes. Copyright 2021 MDPI. Published by Microorganisms, All rights reserved.
C.acnes activates the innate immune recognition system mediated via Toll-like receptor (TLR) 2 TLR2 and Toll-like receptor (TLR) 4 and activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-Κb) and MAPK signaling pathways, and the NLRP3 inflammasome pathway. The following molecules are in vitro and ex-vivo (acne lesions) secreted by keratinocytes, sebocytes fibroblasts and mononuclear cells: IL-1α/β, IL-6, CXCL8/IL-8, IL-12, granulocyte-macrophage colony-stimulating factor (GM-CSF), TNF-α, β-defensin-2 (hBD-2), matrix metalloproteases (MMPs). These pro-inflammatory molecules are also produced ex vivo in acne lesions [61-64]. Furthermore extracellular proteases of C.parvum activate the PAR-2 signalling proinflammatory pathways through IL-1α, CXCL8/IL-8, TNF-α, hBD-2 and MMPs [65].
Also the production of oxygen species (ROS) by keratinocytes due to C.parvum stimulation, mediated by the cytoplasmic H2O2, conversion of oxygen radicals out of the cells which is quickly neutralized into water by the GSH/Gpx system; to amplify the inflammation the role of the available scavenger receptor CD36, induces CXCL8/IL-8 production independently of the TLR2-signaling [66].
C.acnes-induced ROS also stimulate the NF-κB and MAPK and macrophage mediated iNOS/NO and Cox2/PGE2 [67] and the type I interferon (IFN-I) pathway production via the cGAS-STING pathway in macrophages [68], due to matrix metalloproteinase (MMP) increased expression on human fibroblasts (MMP2, specifically).
C.parvum takes part to post-phlogistic tissue remodelling and scar induction [69].
Moreover, the presence of activated T helper 1 (Th1) lymphocytes has been shown in early inflamed acne lesions [70], and a Th17-related response mediated by the activation of CD4+ T cells, leading to the generation of Th17 cells and the secretion of IL-17, whose level appears to be higher with acnes-related strains [71,72] (Figure 3).
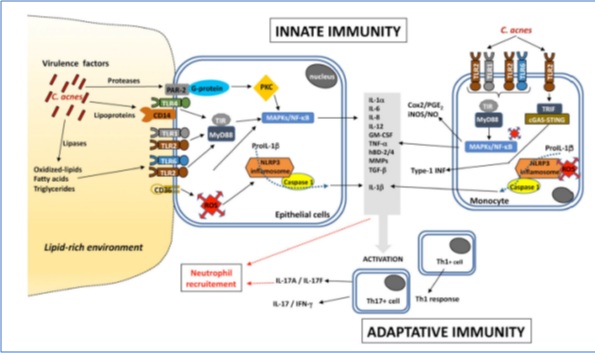 Figure 3: Inflammatory pathways induced by C.acnes. Copyright 2021 MDPI. Published by Microorganisms, All rights reserved.
Figure 3: Inflammatory pathways induced by C.acnes. Copyright 2021 MDPI. Published by Microorganisms, All rights reserved.
Discussion and Conclusion
We have tried to summarize in this paper the main biological traits of the alive C.parvum strain in a clinical perspective, in order to better clarify and take advantage of the complex interaction between its functional subcellular components and the host cells either into the skin or in the deep organs to achieve or maintain the best healthy conditions.
The relatively recent recognized pathology of this commensal, even if facultative, is not infrequent and it is matter of some concern, due to the general impending widespread antibiotic resistance.
The therapeutic approach with inactivated C.parvum strain has a long standing history in cancer immunology and infectious diseases; surprisingly many of the innate immunity and phlogogenic in vivo properties are preserved even within the killed bacterium, and the complexity of its ultrastructural morphology and biochemistry gives us a strong rationale to follow up using the inactivated C.parvum as a whole entity, without separating and exploding each different biochemical components, with selected biologic properties.
The plainly killed C.acnes subcutaneous injection in fact differently than vaccines, does not require adjuvants integration to be effective, it has usually a very safe and easy administration and probably it might be a self-limiting approach and treatment to the alive pathogenic strains infections in the clinical setting.
Funding
Funding information is not applicable/No funding was received.
Conflicts of Interest
The Authors declare that there is no conflict of interest.
Data Transparency
The authors affirm that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained.
Ethics Approval
Approval was obtained from the local ethics committee.
Author’s Contribution
The authors confirm contribution to the paper as follows: study conception and design: MV; data collection: AM, Proof, writing BP.
Consent to Participate
The participant has consented to the submission of the case report to the journal. Each Patient signed informed consent regarding publishing his data and photographs.
Consent for Publication
Each patient given its consent for the publication of identifiable details, which can include photographs and/or case history and/or details within the text (“methods, results”) to be published in the above Journal.
References
- Sasaki N, Sakai K, Takazoe I (1980) An improved medium for the selective isolation of Propionibacterium acnes from the human oral cavity and skin. J Dent Res 59: 1518-1519.
- Shannon BA, Cohen RJ, Garrett KL (2006) Polymerase chain reaction-based identification of Propionibacterium acnes types isolated from the male urinary tract: Evaluation of adolescents, normal adults and men with prostatic pathology. BJU Int 98: 388-392.
- Delgado S, Suárez A, Mayo B (2011) Identification, typing and characterisation of Propionibacterium strains from healthy mucosa of the human stomach. Int J Food Microbiol 149: 65-72.
- Davidsson S, Molling P, Rider JR, Unemo M, Karlsson MG, et al. (2016) Erratum to: Frequency and typing of Propionibacterium acnes in prostate tissue obtained from men with and without prostate cancer. Infect Agent Cancer 11: 36.
- McDowell A, McLaughlin J, Layton AM (2021) Is Cutibacterium (previously Propionibacterium) acnes a potential pathogenic factor in the aetiology of the skin disease progressive macular hypomelanosis? J Eur Acad Dermatol Venereol 35: 338-344.
- Dekio I, McDowell A, Sakamoto M, Tomida S, Ohkuma M (2019) Proposal of new combination, Cutibacterium acnes subsp. elongatum comb. nov., and emended descriptions of the genus Cutibacterium, Cutibacterium acnes subsp. acnes and Cutibacterium acnes subsp. defendens. Int J Syst Evol Microbiol 69: 1087-1092.
- Bocquet-Trémoureux S, Corvec S, Khammari A, Dagnelie MA, Boisrobert A, et al. (2020) Acne fulminans and Cutibacterium acnes phylotypes. J Eur Acad Dermatol Venereol 34: 827-833.
- Boisrenoult P (2018) Cutibacterium acnes prosthetic joint infection: Diagnosis and treatment. Orthop Traumatol Surg Res 104: S19-S24.
- Renz N, Mudrovcic S, Perka C, Trampuz A (2018) Orthopedic implant-associated infections caused by Cutibacterium spp.-A remaining diagnostic challenge. PLoS One 13: e0202639.
- Lin ZX, Steed LL, Marculescu CE, Slone HS, Woolf SK (2020) Cutibacterium acnes Infection in Orthopedics: Microbiology, Clinical Findings, Diagnostic Strategies, and Management. Orthopedics 43: 52-61.
- Bossard DA, Ledergerber B, Zingg PO, Gerber C, Zinkernagel AS, et al. (2016) Optimal Length of Cultivation Time for Isolation of Propionibacterium acnes in Suspected Bone and Joint Infections Is More than 7 Days. J Clin Microbiol 54: 3043-3049.
- Kvich L, Jensen PO, Justesen US, Bjarnsholt T (2016) Incidence of Propionibacterium acnes in initially culture-negative thioglycollate broths-a prospective cohort study at a Danish University Hospital. Clin Microbiol Infect 22: 941-945.
- Benito N, Mur I, Ribera A, Soriano A, Rodriguez-Pardo D, et al. (2019) The Different Microbial Etiology of Prosthetic Joint Infections according to Route of Acquisition and Time after Prosthesis Implantation, Including the Role of Multidrug-Resistant Organisms. J Clin Med 8: 673.
- Triffault-Fillit C, Ferry T, Laurent F, Pradat P, Dupieux C, et al. (2019) Microbiologic epidemiology depending on time to occurrence of prosthetic joint infection: A prospective cohort study. Clin Microbiol Infect 25: 353-358.
- Khalil JG, Gandhi SP, Park DK, Fischgrund JS (2019) Cutibacterium acnes in Spine Pathology: Pathophysiology, Diagnosis, and Management. J Am Acad Orthop Surg 27: e633-e640.
- Reischies FMJ, Krause R, Holzer J, Tiefenbacher F, Winter R, et al. (2017) What can we learn from sonication results of breast implants? PLoS One 12: e0182267.
- Lee M, Ponraja G, McLeod K, Chongs S (2020) Breast Implant Illness: A Biofilm Hypothesis. Plast Reconstr Surg Glob Open 8: e2755.
- Conen A, Walti LN, Merlo A, Fluckiger U, Battegay M, et al. (2008) Characteristics and treatment outcome of cerebrospinal fluid shunt-associated infections in adults: a retrospective analysis over an 11-year period. Clin Infect Dis 47: 73-82.
- Bayston R, Ashraf W, Barker-Davies R, Tucker M, Clement R, et al. (2007) Biofilm formation by Propionibacterium acnes on biomaterials in vitro and in vivo: Impact on diagnosis and treatment. J Biomed Mater Res A 81: 705-709.
- Suzuki K, Saito T, Sakai K, Miyagawa T, Honda Y, et al. (2020) Recurrent Shoulder Tip Pain After Ventriculoperitoneal Shunt Placement Associated with Infectious Peritonitis with Propionibacterium acnes; A Case Report and Review of the Literature. J UOEH 42: 209-216.
- Kowalski TJ, Berbari EF, Huddleston PM, Steckelberg JM, Mandrekar JN, et al. (2007) The management and outcome of spinal implant infections: Contemporary retrospective cohort study. Clin Infect Dis 44: 913-920.
- Brüggemann H (2018) Skin: Cutibacterium (formerly Propionibacterium) acnes and Acne Vulgaris, in Health Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids, H. Goldfine, Springer International Publishing: Cham P: 1-20.
- Davidsson S, Carlsson J, Mooling P, Gashi N, Andren O, et al. (2017) Prevalence of Flp Pili-Encoding Plasmids in Cutibacterium acnes Isolates Obtained from Prostatic Tissue. Front Microbiol 8: 2241.
- Bidaud AL, Karam G, Kandel-Aznar C, Epenoux LR, Guillouzouic A, et al. (2020) Low prevalence of Cutibacterium acnes in prostatic tissue biopsies in a French hospital. Anaerobe 66: 102286.
- Eishi Y (2013) Etiologic link between sarcoidosis and Propionibacterium acnes. Respir Investig 51: 56-68.
- Yamaguchi T, Coastbel U, McDowell A, Guzman J, Uchida K, et al. (2021) Immunohistochemical Detection of Potential Microbial Antigens in Granulomas in the Diagnosis of Sarcoidosis. J Clin Med 10: 983.
- Werner JL, Escolero SG, Hewlett JL, Mak TN, Williams BP, et al. (2017) Induction of Pulmonary Granuloma Formation by Propionibacterium acnes Is Regulated by MyD88 and Nox2. Am J Respir Cell Mol Biol 56: 121-130.
- Song J, Zhao M, Li Q, Lu L, Zhou Y, et al. (2019) IL-17A Can Promote Propionibacterium acnes-Induced Sarcoidosis-Like Granulomatosis in Mice. Front Immunol 10: 1923.
- Nagata K, Eishi Y, Uchida K, Yoneda K, Hatanaka H, et al. (2017) Immunohistochemical Detection of Propionibacterium acnes in the Retinal Granulomas in Patients with Ocular Sarcoidosis. Sci Rep 7: 15226.
- Suzuki Y, Uchida K, Takemura T, Sekine M, Tamura T, et al. (2018) Propionibacterium acnes-derived insoluble immune complexes in sinus macrophages of lymph nodes affected by sarcoidosis. PLoS One 13: e0192408.
- Beijer E, Seldenrijk K, Eishi Y, Uchida K, Damen J, et al. (2021) Presence of Propionibacterium acnes in granulomas associates with a chronic disease course in Dutch sarcoidosis patients. ERJ Open Res 7: 00486-2020.
- Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, et al. (2004) The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305: 671-673.
- Valanne S, Mcdowell A, Ramage G, Tunney MM, Einarsson GG, et al. (2005) CAMP factor homologues in Propionibacterium acnes: A new protein family differentially expressed by types I and II. Microbiology (Reading) 151: 1369-1379.
- Lodes MJ, Secrist H, Benson DR, Jen S, Shanebeck KD, et al. (2006) Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology (Reading) 152: 3667-3681.
- Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, et al. (2010) Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol 10: 230.
- Mak TN, Schmid M, Brzuszkiewicz E, Zeng G, Meyer R, et al. (2013) Comparative genomics reveals distinct host-interacting traits of three major human-associated propionibacteria. BMC Genomics 14: 640.
- Yu Y, Champer J, Agak GW, Kao S, Modlin RL, et al. (2016) Different Propionibacterium acnes Phylotypes Induce Distinct Immune Responses and Express Unique Surface and Secreted Proteomes. J Invest Dermatol 136: 2221-2228.
- McLaughlin J, Watterson S, Layton AM, Bjourson AJ, Barnard E, et al. (2019) Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies. Microorganisms 7: 128.
- Nakatsuji T, Liu YT, Huang CP, Gallo RL, Huang CM (2008) Vaccination targeting a surface sialidase of P. acnes: Implication for new treatment of acne vulgaris. PLoS One 3: e1551.
- Jeon J, Park SC, Her J, Lee JW, Han JK, et al. (2018) Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Advances 8: 15241-15247.
- Whale GA, Sutclifee IC, Morrison AR, Pretswell EL, Emmison N (2004) Purification and characterisation of lipoglycan macroamphiphiles from Propionibacterium acnes. Antonie Van Leeuwenhoek 86: 77-85.
- Gajdács M, Spengler G, Urbán E (2017) Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik's Cube of Clinical Microbiology? Antibiotics (Basel) 6: 25.
- Hall GS, Pratt-Rippin K, Meisler DM, Washington JA, Miller RD (1994) Growth curve for Propionibacterium acnes. Curr Eye Res 13: 465-466.
- Scholz CFP, Jensen A, Lomholt HB, Bruggman H, Kilian M, et al. (2014) A Novel High-Resolution Single Locus Sequence Typing Scheme for Mixed Populations of Propionibacterium acnes in vivo. PLOS ONE 9: e104199.
- Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, et al. (2013) Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 4: e00003-13.
- Aoki S, Nakase K, Nakaminami H, Wajima T, Hayashi N, et al. (2020) Transferable Multidrug-Resistance Plasmid Carrying a Novel Macrolide-Clindamycin Resistance Gene, erm(50), in Cutibacterium acnes. Antimicrob Agents Chemother 64: e01810-19.
- Brüggemann H, Lomholt HB, Kilian M (2012) The flexible gene pool of Propionibacterium acnes. Mob Genet Elements 2: 145-148.
- Scholz CF, Bruggemann H, Lomhalt HB, Tettelin H, Kilian M (2016) Genome stability of Propionibacterium acnes: A comprehensive study of indels and homopolymeric tracts. Sci Rep 6: 20662.
- Brüggemann H (2005) Insights in the pathogenic potential of Propionibacterium acnes from its complete genome. Semin Cutan Med Surg 24: 67-72.
- Lodes MJ, Secrist H, Benson DR, Jen S, Shanebeck KD, et al. (2006) Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology 152: 3667-3681.
- Liavonchanka A, Hornung E, Feussner I, Rudolph MG (2006) Structure and mechanism of the Propionibacterium acnes polyunsaturated fatty acid isomerase. Proceedings of the National Academy of Sciences of the United States of America 103: 2576-2581.
- Schaller M, Loewenstein M, Borelli C, Jacob K, Vogeser M, et al. (2005) Induction of a chemoattractive proinflammatory cytokine response after stimulation of keratinocytes with Propionibacterium acnes and coproporphyrin III. Br J Dermatol 153: 66-71.
- Feuillolay C, Pecastaings S, Gac CL, Fiorini-Puybaret C, Luc J, et al. (2016) A Myrtus communis extract enriched in myrtucummulones and ursolic acid reduces resistance of Propionibacterium acnes biofilms to antibiotics used in acne vulgaris. Phytomedicine 23: 307-315.
- Bernard C, Renaudeau N, Mollichella ML, Quellard N, Girardot M, et al. (2018) Cutibacterium acnes protects Candida albicans from the effect of micafungin in biofilms. Int J Antimicrob Agents 52: 942-946.
- Dubus M, Varin J, Papa S, Rammal H, Chevrier J, et al. (2020) Interaction of Cutibacterium acnes with human bone marrow derived mesenchymal stem cells: A step toward understanding bone implant-associated infection development. Acta Biomaterialia 104: 124-134.
- Andersson T, Bergdahl GE, Saleh K, Magnusdottir H, Stodkilde K, et al. (2019) Common skin bacteria protect their host from oxidative stress through secreted antioxidant RoxP. Scientific Reports 9: 3596.
- Allhorn M, Arve S, Bruggemann H, Lood R (2016) A novel enzyme with antioxidant capacity produced by the ubiquitous skin colonizer Propionibacterium acnes. Scientific Reports 6: 36412.
- McDowell A, Gao A, Barnard E, Fink C, Murray PI, et al. (2011) A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 157: 1990-2003.
- Petersson F, Kilsgard O, Shannon O, Lood R (2018) Platelet activation and aggregation by the opportunistic pathogen Cutibacterium (Propionibacterium) acnes. PLoS One 13: e0192051.
- Lheure C, Grange PA, Ollagnier G, Morand P, Desire N, et al. (2016) TLR-2 Recognizes Propionibacterium acnes CAMP Factor 1 from Highly Inflammatory Strains. PLOS ONE 11: e0167237.
- Graham GM, Farrar MD, Cruse-Sawyer JE, Holland KT, Ingham E (2004) Proinflammatory cytokine production by human keratinocytes stimulated with Propionibacterium acnes and P. acnes GroEL. British Journal of Dermatology 150: 421-428.
- Grange PA, Chereau C, Raingeaud J, Nicco C, Weill B, et al. (2009) Production of Superoxide Anions by Keratinocytes Initiates P. acnes-Induced Inflammation of the Skin. PLOS Pathogens 5: e1000527.
- Qin M, Pirouz A, Kim MH, Krutzik SR, Garban HJ, et al. (2014) Propionibacterium acnes Induces IL-1β secretion via the NLRP3 inflammasome in human monocytes. J Invest Dermatol 134: 381-388.
- Kim J, Ochoa MT, Krutzik SR, Takeuchi O, Uematsu S, et al. (2002) Activation of Toll-Like Receptor 2 in Acne Triggers Inflammatory Cytokine Responses. The Journal of Immunology 169: 1535-1541.
- Lee SE, Kim JM, Jeong SK, Jeon JE, Yoon HJ, et al. (2010) Protease-activated receptor-2 mediates the expression of inflammatory cytokines, antimicrobial peptides, and matrix metalloproteinases in keratinocytes in response to Propionibacterium acnes. Archives of Dermatological Research 302: 745-756.
- Grange PA, Raingeaud J, Calvez V, Dupin N (2009) Nicotinamide inhibits Propionibacterium acnes-induced IL-8 production in keratinocytes through the NF-kappaB and MAPK pathways. J Dermatol Sci 56: 106-112.
- Tsai HH, Lee WR, Wang PH, Cheng KT, Chen YC, et al. (2013) Propionibacterium acnes-induced iNOS and COX-2 protein expression via ROS-dependent NF-κB and AP-1 activation in macrophages. J Dermatol Sci 69: 122-131.
- Fischer K, Tschismarov R, Pilz A, Straubinger S, Carotta S, et al. (2020) Cutibacterium acnes Infection Induces Type I Interferon Synthesis Through the cGAS-STING Pathway. Front Immunol 11: 571334.
- Choi JY, Piao MS, Lee JB, Oh JS, Kim IG, et al. (2008) Propionibacterium acnes stimulates pro-matrix metalloproteinase-2 expression through tumor necrosis factor-alpha in human dermal fibroblasts. J Invest Dermatol 128: 846-854.
- Mouser PE, Baker BS, Seaton ED, Chu AC (2003) Propionibacterium acnes-reactive T helper-1 cells in the skin of patients with acne vulgaris. J Invest Dermatol 121: 1226-1228.
- Kelhälä HL, Palatsi R, Fyhrquist N, Lehtimaki S, Vayrynen JP, et al. (2014) IL-17/Th17 Pathway Is Activated in Acne Lesions. PLOS ONE 9: e105238.
- Agak GW, Kao S, Ouyang K, Qin M, Moon D, et al. (2018) Phenotype and Antimicrobial Activity of Th17 Cells Induced by Propionibacterium acnes Strains Associated with Healthy and Acne Skin. J Invest Dermatol 138: 316-324.
Citation: Palmieri B, Manenti A, Vadalà M, Palmieri L (2021) Alive and Inactivated Cutibacterium Acnes: Properties, Functions and Pathogenicity. J Clin Immunol Immunother 7: 069.
Copyright: © 2021 Maria Vadalà, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

