
Ribosome Profiling in Streptococcus pneumoniae Reveals the Role of Methylation of 23S rRNA Nucleotide G748 on Ribosome Stalling
*Corresponding Author(s):
Tatsuma ShojiLaboratory Of Microbiology And Immunology, Graduate School Of Pharmaceutical Sciences, Chiba University, Japan
Tel:+81 8051809605,
Email:tatsumashoji@bioinforest.com
Abstract
Background: Many nucleotides in 23S rRNA are methylated post-transcriptional by methyltransferases and cluster around the Peptidyl Transferase Center (PTC) and the Nascent Peptidyl Exit Tunnel (NPET) located in 50S subunit of 70S ribosome. Biochemical interactions between a nascent peptide and the tunnel may stall ribosome movement and affect expression levels of the protein. However, no studies have shown a role for NPET on ribosome stalling using an NPET mutant.
Results: A ribosome profiling assay in Streptococcus pneumoniae demonstrates for the first time that an NPET mutant exhibits completely different ribosome occupancy compared to wild-type. We demonstrate, using RNA foot printing that changes in ribosome occupancy correlate with changes in ribosome stalling. Further, statistical analysis shows that short peptide sequences that cause ribosome stalling are species-specific and evolutionarily selected. PET structure is required to realize these specie-specific ribosome stalling.
Conclusion: Results support the role of NPET on ribosome stalling. NPET structure is required to realize the species-specific and evolutionary conserved ribosome stalling. These findings clarify the role of NPET structure on the translation process.
Keywords
Ribosome stalling; Ribosome profiling; Ribosomopathy; 23S rRNA modification; Streptococcus pneumoniae
Background
Endogenous rRNA modifying enzymes methylate or pseudouridylate specific rRNA nucleotides at functionally important regions in the ribosome, such as the Peptidyl Transferase Center (PTC) [1]. Approximately one-third of modified residues of 23S rRNA are clustered around the Nascent Peptide Exit Tunnel (NPET) [2].While the role of rRNA modification remains unclear, though it is generally believed that they have fine-tune functions of the ribosome in translation [3], especially under the stress conditions [4], via the biochemical interactions between the nascent peptide and tunnel [5,6]. These interactions may stall ribosome movement and thus affect the expression level of the protein [6]. However, no studies have shown the role of NPET in ribosome stalling using an NPET mutant.
Some modifications of NPET are important for determining antibiotic resistance or susceptibility [7,8]. While the role of the methylation at G748 (m1G748) in Streptococcus pneumoniae remains unclear, we previously showed that inactivation of the methyltransferase RlmAII, which methylates the N-1 position of nucleotide G748 located near the PTC, results in increased resistance to telithromycin (TEL) in erm (B)-carrying S .pneumoniae [7].
We explored the role of NPET structure in translation initially by establishing the ribosome profiling assay in S. pneumonia and investigating ribosomal distribution in both wild-type and RlmAII-deficient S. pneumoniae. Subsequent analysis showed that m1G748 is responsible for ribosome stalling and plays a role in species specificity.
Results
The loss of N-1 methylation at G748 greatly changes the distribution of ribosome’s
We constructed two S. pneumonia mutant strains, Sp284 and Sp379 to investigate the role of NPET. Sp284 is a RlmAII-disrupted mutant harboring pTKY1111 encoding tlr B of the S1 strain [7]. Sp379 is an RlmAII-disrupted mutant harboring pTKY1127 encoding tlrB of Sp44 [7]. The latter strains display no methyltransferase activity because of the C23R mutation [7]. The difference between Sp284 and Sp379 is only the capability to methylate G748.
We also performed a deep sequencing-based ribosome profiling analysis in Sp284 and Sp379. Ribosome profiling captures a global snapshot of ribosome positioning and density on template mRNAs with single-nucleotide resolution. The erm (B) operon, where ribosomes stall at the ermBL region [9], was selected, for example, to examine the distribution of ribosomes (Figure 1A). Ribosome position and density in the erm(B) operon in Sp379 was completely different from positions and density in Sp284 (Figure 1A).
The difference in the ribosome occupancy between two strains in erm (B) operon led us to speculate a general role of m1G748 in the translation process. Ribosome density across all ORFs in Sp379 was higher than in Sp284, especially around the latter region, indicating an important role of m1G748 in the translation process, with differences in ribosome position and density across all Open Reading Frames (ORFs) being assessed using a metagene analysis (Figure 1B).
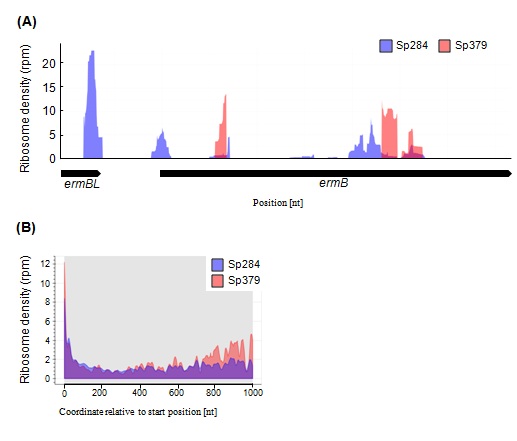 Figure 1: Differences in ribosome density (RD) profiles between Sp284 and Sp379.
Figure 1: Differences in ribosome density (RD) profiles between Sp284 and Sp379.
(A) RD in erm(B) in Sp284 or Sp379. (B) Metagene profile of RD in Sp284 or Sp379. RD across all ORFs was aligned relative to the start position.
m1G748 affects ribosome stalling
A high number of Ribosome-Protected Footprints (RPFs) mappings to the transcriptome (mRNA-seq) at a unique position are indicative of ribosome stalling [10]. Thus, observed global changes implya role for m1G748 in ribosome stalling.
We tested this hypothesis by first constructing erm(B) operon-over expression s. pneumoniae strains Sp380 and Sp382 from Sp284 and Sp379 respectively, to clarify differences between Sp284 and Sp379 (Tables 1 and 2).We then probed ribosomes of Sp380 and Sp382 before and after extracting total RNA with dimethyl sulfate.
|
Strain |
Relevant characteristics |
Reference of source |
|
Streptococcus pneumoniae |
||
|
S1 |
TEL resistance clinical isolate |
[7] |
|
Sp36 |
TEL-resistant mutants of S1 isolated from 8 μg/mL TEL-containing BHI-Y agar plates |
[7] |
|
Sp274 |
ΔtlrB::aad(9) in S1 |
[7] |
|
Sp284 |
Sp274 harboring pTKY1111 |
This study |
|
Sp379 |
Sp274 harboring pTKY1127 |
This study |
|
Sp380 |
S1 harboring pTKY1041 |
This study |
|
Sp382 |
Sp274 harboring pTKY1041 |
This study |
Table 1: Bacterial strains.
|
Plasmid |
Relevant characteristics |
Reference of source |
|
pUC18 |
Cloning vector |
Lab. collection |
|
pLZ12-Km2 |
Shuttle vector |
[11] |
|
pTKY862 |
pLZ12-Km2 with Sp resistant cassette ad(9) |
[12] |
|
pTKY1041 |
pLZ12-Km2 with 1401 bperm(B) fragment from S1 |
[7] |
|
pTKY1109 |
pUC18 with 328 bptlrB fragment |
[7] |
|
pTKY1110 |
pUC18 with disrupted tlrB fragment by insertion of an Sp resistant cassette aad(9) |
[7] |
|
pTKY1111 |
pLZ12-Km2 with 1065 bptlrB fragment from S1 |
[7] |
|
pTKY1127 |
pLZ12-Km2 with 1065 bptlrB fragment from Sp44 |
[7] |
Table 2: Plasmids.
Figure 2A shows the secondary mRNA structure from the ermBL region that was previously demonstrated [9]. Consistent with this report, nucleotides in the stems were protected from chemical modification when probing after extracting total RNA in both Sp380 and Sp382 strains. Conversely, these regions were not protected when probing before extracting total RNA in Sp380 (Figure 2B), but these regions remained unprotected in Sp382 (Figure 2B). Stems were likely disrupted in Sp380 in vivo presumably due to ribosome stalling in the ermBL region. m1G748 affects such stalling.
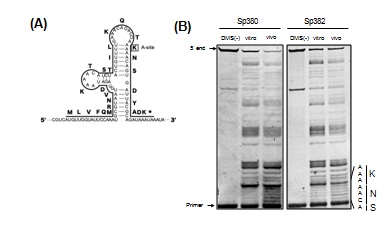
Figure 2: RNA foot printing assay for the ermBL region.
(A) The secondary structure model of ermBL region in erm(B) mRNA suggested in [9]. Amino acids in at the A-site of stalled ribosomes in the ermBL region are boxed. (B) RNA foot printing of the ermBL region indicates a difference in ribosome stalling between Sp380 and Sp382. DMS was added to bacterial cultures (in vivo) or the total RNAs (in vitro). Reactions were quenched, and total RNA was purified and used in primer extension assays to detect base modifications. Unmodified RNA was used as a control (DMS (-)). The gel shows primer extension products from bases 117 to 152. The sequence around the calculated P-site and encoded amino acids are indicated on the side of the gel.
Characterization of effects of m1G748 on ribosome stalling
Ribosome stalling does not always mean an end to translation. Translation resumes in some cases [13,14]. Such cases are termed “transient stalling” [15] and may contribute to co translational protein and be evolutionarily preferred [16,17]. In contrast, "strong stalling" does not appear to restart and requires rescue [18,19]. We examined the type of ribosome stalling affected by m1G748 byfirst defining A-site peaks (see Methods for details) and counted the number of A-site peaks across all CDSs (Figures 3A and 3B). Two populations observed in Sp284 (Figure 3A) were consistent with a previous report [18]. However, surprisingly, almost no transient stalling was observed in Sp379 (Figure 3B), indicating a role for m1G748 for keeping transient stalling. A slight decrease in the number of peaks of strong stalling was observed compared to Sp284 (Figures 3A and 3B).
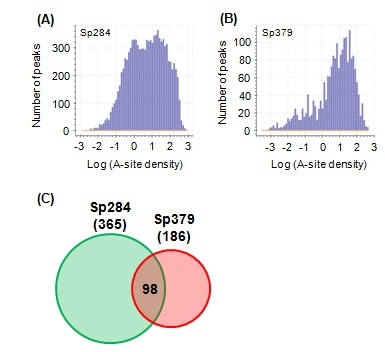 Figure 3: The effect of m1G748 on transient stalling and strong stalling.
Figure 3: The effect of m1G748 on transient stalling and strong stalling.
(A,B) Histograms showing the distribution of A-site density in Sp284 and Sp379, respectively. (C) Venn diagram for stalling peptide sequences in Sp284 and Sp379. The number of stalling peptide sequences is shown in parentheses. The number in the intersection region reflects common stalling peptides.
Stalling peptides were identified as previously described [15] to further examine the role of m1G748 on strong stalling. Briefly, we defined strong stalling as described in Methods and collected nascent peptide sequences in the exit tunnel for strong stalling events. We then calculated the probability of occurrence for 8,000 tripeptides and defined stalling peptides as tripeptides with a probability higher than 0.9999. Surprisingly, few stalling peptides were common between Sp284 and Sp379 (Figure 3C), suggesting that the position of “strong stalling” is different between the two strains.
Several known stalling peptides, such as PPP, KKK and KKR, are common among some organisms, including Saccharomyces cerevisiae and Escherichia coli [20,21]. However, stalling peptides in Sp284 and Sp379 did not include these previously reported molecules [see Additional file 1], indicating that ribosome stalling is species-specific.
m1G748 is required to realize evolutionarily conserved ribosome stalling
Stalling peptides in S. pneumonia were different in previous reports, which led us to speculate on how they are distributed in the proteome. We examined relationships between stalling peptides and proteome in S. pneumoniae, by initially identifying 360 of over-represented and 382 under-represented examples for three amino acids in the S. pneumoniae proteome as previously [15]. Enrichment of over-and under-represented tripeptides in stalling peptides was investigated using Fisher's exact test (Figure 4). Over-represented peptides were significantly enriched in the set from Sp379, with under-represented peptides being significantly enriched in the stalling peptide set of Sp284, indicating that the strong stalling is not evolutionarily favored [see Additional file 2].m1G748 is likely required to realize evolutionarily conserved ribosome stalling.
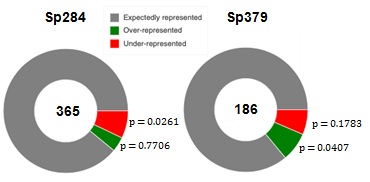 Figure 4: Enrichment of over- and under-represented stalling peptides.
Figure 4: Enrichment of over- and under-represented stalling peptides.
The number of the stalling peptides is represented at the center of the donut. The p-value represents statistical significance of the over- and under-represented fractions.
Discussion
We investigated the role of NPET in translation using a ribosome profiling assay in S. pneumoniae. The finding that the loss of the methyl group at m1G748 had a notable impact on the distribution of the ribosomes and ribosome stalling (Figures 1 and 2) highlights the importance of the NPET structure and explains why rRNA modifications are clustered near the PTC and NPET. The role of NPET using other NPET mutants will be investigated in the future.
Alteration of NPET results in changes to the stalling peptide set (Figure 3C). Thus, translation depends not only on mRNA sequence but also on the structure of NPET. Prediction of ribosome stalling based only on mRNA sequence would be difficult. Using NPET information is crucial because the structure of the NPET is not necessarily unique in a cell [22], with some studies having tried to predict ribosome occupancy or stalling [21,23], and performance of developed software could be improved with additional parameters for NPET structure.
A global change in stalling position may result in a global effect on cell proteome. This effect might explain why RlmAII mutants of S. pnuemoniae are rarely clinically isolated [24]. The loss of the m1G748 methyl group inhibits binding of telithromycin to the exit tunnel [7] and may also cause a decrease in fitness of S. pneumonia in a clinical setting. This concept might be useful since antibiotics that compromise targets that maintain healthy ribosomes would not cause resistant bacteria.TEL-resistant S. pneumoniae ribosomes are unhealthy.
The ribosomopathy encompasses diseases caused by abnormalities in the structure or function of ribosomal proteins or rRNA genes or other genes whose products are involved in ribosome biogenesis [25-27]. Skeletal muscle atrophy [28], Diamond–Blackfan anemia [26] and Treacher Collins syndrome [26] are examples of ribosomopathy. However, no reports regarding exit tunnel-induced ribosomopathy exist. The present study examined S. pneumoniae; however, exit tunnel-derived ribosomopathy might be common among organisms, including humans. For example, ribosomal protein L17 (RPL17) is up regulated in parallel with stress vulnerability [29]. RPL17 is located near the end of the exit tunnel [22]. Therefore, RPL17 could be responsible for ribosome stalling. This concept, exit-tunnel-induced ribosomopathy, might explain the mechanism of the disorder in the future.
Conclusion
We demonstrate the role of m1G748 on ribosome stalling in S. pneumoniae. m1G748 is required to realize species-specific and evolutionarily conserved stalling. The loss of the methyl group at m1G748 has a great impact on the distribution of the ribosomes and ribosome stalling, and results in exit-tunnel-induced ribosomopathy. This might be the reason why RlmAII mutants of S. pnuemoniae are rarely clinically isolated. This study is the first to show the role of NPET using an NPET mutant. These findings clarify the role of NPET structure on the translation process.
Methods
Bacterial strains, plasmids, and media
Bacterial strains and plasmids are shown in tables 1 and 2, respectively. S. pneumoniae strain S1 with reduced TEL susceptibility (MIC, 2μg/ml) was clinically isolated in Japan [7]. Pneumococci were routinely cultured at 37°Cand 5% CO2 in air in a brain-heart infusion with 0.5% yeast extract (BHI-Y) broth and BHI-Y agar, supplemented with 5% horse blood. E. coli was grown in L broth (1% bact-tryptone, 0.5% bact yeast extract, 0.5% sodium chloride, pH 7.4) and L agar. When necessary, medium was supplemented with kanamycin (25–500μg/mL), spectinomycin (100 μg/mL) and ampicillin (25 μg/mL).
Transformation
Synthetic Competence-Stimulating Peptide (CSP) 1 and the method of Iannelli and Pozzi [30] were used to transform S. pneumoniae S1 into a transformation-competent state.
RNA-Seq
S.pneumoniae cultures were grown to log-phase; 2.8 mL of cultures were added to 2.8 mL of 100°C preheated RNA lysis buffer (1% SDS, 0.1 M NaCl and 8mM EDTA) and vortexed for 2min. The resulting lysates were added to 5.6 mL of 100°C preheated acid phenol (Sigma-Aldrich) and vortexed for 5 min. After centrifuging, RNA was extracted from the aqueous phase using DirectZol (Zymo Research). rRNA was removed from total RNA using MICROB Express (Ambion). Resulting total mRNA (400ng as an input) was used for constructing the DNA library, using KAPA Stranded RNA-Seq Library Preparation Kit Illumina platforms (KK8400). DNA libraries were sequenced using the Illumina HiSeq 1500 system and single-end reads. Illumina libraries were preprocessed by clipping the Illumina adapter sequence using Trimmomatic v.0.39 [31] and then aligned to the S1 genome sequence [7] using HISAT2 v.2.2.1 [32].
Ribo-Seq
Libraries were prepared as previously described with some modifications (see below) [10].
Cell growth and harvest: S. pneumoniae cultures (2.4L) were grown to log-phase. Cells were pretreated for 2 min with ∼100 μg/mL chloramphenicol. Immediately after chloramphenicol pretreatment, cultures were placed on ice. Cells were pelleted by centrifugation at 8,000 ×g for 15 min at 4°C. After decanting the supernatant, cell pellets were resuspended in 2.5 mL of prechilled resuspension buffer [10mM MgCl2, 100mM NH4Cl, 20mM Tris (pH 8.0), and 1mM chloramphenicol].
Lysate preparation and Nuclease digestion: Cells were sonicated on ice and centrifuged at 12,000 × g for 10 min at 4°C. Aliquots of lysate, containing 25 Abs260 ribosome units (1 A260 = 12 μg/μL) [5] were digested with 60 U of MNase (Roche) and 60 U of SUPERase. In (Ambion) with the addition of chloramphenicol to a final concentration of 1mM to remove unprotected mRNA and generate footprint fragments. Digestion reactions were incubated for 1 h at 25°C and quenched with the addition of EGTA to a final concentration of 6 mM.
Sucrose fractionation: Linear sucrose gradients [5-40% (wt/vol)] were prepared with 7.6mL of buffer A and buffer B [10mM MgCl2, 100mM NH4Cl, 2mM DTT, 20mM, 0.2 mMchloramphenicol, Tris pH 7.8 and 5% or 40% Sucrose, respectively], by loading Buffer B on Buffer A in 16PA tube and placed vertically for 12 hr at 4°C the placed horizontally for 2 hr at 4°C.
Digested samples were carefully loaded onto prepared gradients and centrifuged at 124,700 × g for 8 hr at 4°C in a P28S2 rotor. Sucrose gradients were fractionated manually into 200 μL portions and A260was recorded for each fraction. Absorbance values were graphed in Microsoft Excel to determine ribosome footprint associated fractions (RPF fractions). RPF were consequently pooled.
Ribosome footprint preparation: RNA was purified from RPF using the SDS/hot acid phenol. Three mL of samples were first denatured with SDS to a final concentration of 1% (wt/vol) and 2.7mL of preheated acid phenol (65°C) (Sigma-Aldrich). Mixtures were vortexed for 5 min at 65°C. After centrifuging, aqueous phases were mixed with one volume of acid phenol, and 0.9 volumes of chloroform/isoamyl alcohol (24:1). RNA was precipitated with ethanol and resuspended in 10 μL of 10mM Tris (pH 8.0).
Ribosome footprint samples were resolved on denaturing polyacrylamide gels for size selection of footprint fragments. RNA samples were prepared for electrophoresis by adding 2x Novex TBE-Urea Sample Buffer (Invitrogen). Ladder standards used a 0.05 μg/μL 10-bp DNA Ladder (Invitrogen), prepared in 2x Novex TBE-Urea Sample Buffer and 10mM Tris (pH 8.0). Samples were resolved on a 15% TBE-Urea gel in 1 × TBE buffer for 65 min at 200V. Gels were stained for 3 min with SYBR Gold Nucleic Acid Gel Stain (diluted from 10,000x in 1x TE; Invitrogen) and visualized by UV transillumination. A band between 20 and 45 was excised using the 10-bp DNA ladder to identify footprint fragments. RNA was recovered using the ZR small-RNA PAGE Recovery Kit (Zymo Research) following the manufacturer's protocol, except that RNA was eluted from the final spin column with 15 μL of 10mM Tris (pH 8.0). Collected RNA was quantified and characterized by using a small-RNA chip on the Agilent BioAnalyzer (Agilent Technologies).
Dephosphorylation: Three hundred pmol of footprints were denatured for 2min at 80°C and placed on ice. The 3' ends were dephosphorylated with T4 polynucleotide kinase (T4 PNK; NEB) in the following reaction mix: 1 × T4 PNK reaction buffer (without ATP), 20 U SUPERase. In and 10 U T4 PNK. Reactions were incubated at 37°C for 1 hr. The enzyme was then heat-inactivated for 10 min at 75°C. RNA was precipitated with isopropanol and resuspended in 10 μL of 10mM Tris (pH 8.0).
Linker-1 Ligation: Twenty pmolof dephosphorylated RNA was prepared by diluting with 10mM Tris (pH 8.0). One μg of Linker-1 (5'-App CTGTAGGCACCATCAAT ddC-d') was added to RNA samples. Mixtures were denatured for 90 s at 80°C, then cooled to room temperature for 15 min. Ligation of RNA to Linker-1 used the following reaction components: 20% (wt/col) PEG, 10% DMSO, 1 × T4 Ligase reaction buffer, 20U SUPERase • In, and 10 U T4 Ligase 2, truncated (NEB). Reaction mixtures were incubated at 37°C for 1 h. 2 × TBE-Urea sample buffer was added to reaction mixtures. Samples were resolved on a 10% TBE-Urea gel in 1 × TBE buffer at 200V. Gels were stained for 3min in SYBR Gold Nucleic Acid Gel Stain and visualized by UV transillumination. A band between 30 and 70 nt was excised using the 10-bp DNA ladder to mark ligated product size. Ligated RNA was recovered using the ZR small-RNA PAGE Recovery kit. Ligated products were eluted from the final spin column with 6 μL of 10mM Tris (pH 8.0).
Phosphorylation: The collected 3' ligated samples were incubated for 2 min at 80°C and placed on ice. The 5' ends were phosphorylated by T4 PNK in the following reaction mix: 1 × T4 PNK Reaction Buffer, 20 U SUPERase. In 10 U T4 PNK and 1mM ATP. Reactions were incubated at 37°C for 1 hr. The enzyme was heat-inactivated for 10 min at 75°C. RNA was precipitated with isopropanol and resuspended in 6 μL of 10mM Tris (pH 8.0).
Linker-2 ligation: 1 μL of 100 μM Linker-2 (5'-GAGTCTGCGTGTGATTCGGGTTAGGTGTTGGGTTGGGCCA-3') was added to phosphorylate RNA samples. Mixtures were denatured for 15 min at 65°C and placed on ice. Ligation of RNA to Linker-2 used the following reaction mix: 17.5% (wt/vol) PEG, 1 × T4 ligase reaction buffer, 20U SUPERase. In, and 10 U T4 RNA Ligase1 (NEB). Reaction mixtures were incubated at 37°C for 2.5 h. Two × TBE-Urea sample buffer was added and ligated RNA resolved on 10% TBE-Urea gels in 1 × TBE buffer at 200V. Gels were stained for 3min in SYBR Gold Nucleic Acid Gel Stain and visualized by UV transillumination. A band between 90 and 120 nt was excised using the 10-bp DNA ladder to identify ligated products. Ligated RNA was recovered using the ZR small-RNA PAGE Recovery kit. Products were eluted from the final spin column with 6 μL of 10mM Tris (pH 8.0).
Reverse transcription: Four point five μL of ligated samples were mixed with 1 μL of 0.1 μM Linker-1-RT (5'-ATTGATGGTGCCTACAG-3') and 1 μL of 0.5mM dNTP. The resulting mixtures were denatured for 2 min at 80°C then quickly cooled on ice. Samples were incubated at room temperature for 10 min reverse transcription using Superscript III Reverse Transcriptase (Invitrogen) with the following reaction mix: 1 × first strand buffer, 5mMDTT, 20 U SUPERase. In and 200 U Superscript III Reverse Transcriptase. Reaction mixtures (10 μL) were incubated for 1hr at 47°C. RNA products were hydrolyzed by adding 1mMNaOH to a final concentration of 0.1 mM and incubated for 15 min at 95°C. cDNA products were resolved from the unextended primer on a 10% TBE-Urea gel in 1 × TBE buffer at 200V. Samples were prepared for electrophoresis by adding 2 × TBE-Urea sample buffers and denaturing for 5 min at 95°C. Gels were stained for 3 min in SYBR Gold Nucleic Acid Gel Stain and visualized by UV transillumination. A band between 90 and 120 nt was excised using the 10-bp DNA ladder to identify reverse transcription products. DNA was recovered using the ZR small-RNA PAGE Recovery Kit. cDNA products were eluted from the final spin column with 6 μL of 10mM Tris (pH 8.0).
2nd strand DNA synthesis: cDNA (100 pg) was amplified with Q5 High Fidelity Polymerase (NEB) using LInker-2-partial (5'-TTAGGTGTTGGGTTGGGCCA-3') and Linker-1 as primers. Amplified PCR products were purified using AMPure Bead (Beackman Coulter). Double-stranded DNA was eluted from beads with 10 μL of 10mM Tris (pH 8.0).
Library preparation and sequencing: KAPA Hyper Prep Kits from Illumina (KK8500) were used to construct the library with a slight modification to the manufacturer's protocol. The manufacturer's protocol followed the EndRepair and A-tailing steps, where, briefly, 5 μL of KAPA frag buffer was added to 45 μL of purified double-stranded DNA. The resulting DNA library was quantified and characterized using the high-sensitivity DNA chip on an Agilent BioAnalyzer (Agilent Technologies). Libraries were sequenced using an Illumina HiSeq 1500 system and single-end reads after adding PhiX (Illumina) to a final concentration of 30% (vol/vol) to improve sequencing quality.
Sequence analysis: Illumina libraries were preprocessed by clipping adapter sequence (Linker-1 and Linker-2) using TrimmomaticPE v.0.39 [31] for the Illumina adapters and CutAdapt v.2.10 [33] for linker sequences. Sequencing reads were aligned to the S1 genome sequence [7] using HISAT2 v.2.2.1 [32]. The S1 gene feature file [7] was used to identify the CDS region. Sequencing data were deposited in the DDBJ database with accession number DRA011224.
Definition of the ribosome density
The Ribosome Density (RD) was calculated as previously described [34], except that ribosome footprints between 24 and 30 nt were selected for calculations. This range was chosen to assess as many footprints as possible to improve statistical power and to exclude fragments suspected not to represent footprints (see Additional file 3 for the detail).
Metagene analysis
Each normalized RD profile was aligned by its start cod on and averaged across each position, with RD profiles first being scaled by their own mean density to obtain metagene profiles (Figure 1B).
DMS modification of ribosomes
Dimethyl Sulfate (DMS) modification was based on a previously published protocol [7]. S. pneumoniae cultures (2.4L) were grown to log-phase. Cells were pelleted by centrifugation at 8,000 × g for 15 min at 4°C and washed and resuspended in 250 μL of DMS buffer [10mM MgCl2, 50mM sodium cacodylate pH 7.0]. For the modification in vivo, DMS solution was added to a final concentration of 50mM and incubated for 1hr at 20°C. The reaction was quenched by the addition of 62.5 μL ice-cold stop buffer [1M β-mercaptoethanol and 1.5M sodium acetate, pH 7.0]. RNA was extracted using SDS/hot acid phenol method. Preheated (100°C) acid phenol (0.9 volumes) was added to the quenched mixture and vortexed for 5 min. After centrifugation, RNA was purified from the aqueous phase using DirectZol (Zymo Research). For the modification in vitro, RNA was first extracted from the cell suspension using the same method as for in vivo. After purifying the total RNA, DMS solution was added to a final concentration of 50mM in DMS buffer and incubated for 1hr at 20°C then quenched. RNA was precipitated with ethanol and resuspended in 20 μL of 10 mM Tris (pH 8.0).
Primer extension
The degree of methylation of each RNA was assayed by primer extension initially described by Morgan et al., [35]. Briefly, 210 μg of DMS-probed total RNA, 400 nM concentration of an oligonucleotide primer containing 5' - linked fluoresce in isothiocyanate (FITC) (5'-[FITC]GAATTAATCTAACGTATTTATTTATCTGCGTAATCA -3'), and 500 μM dNTP were mixed, heated to 95°C for 1 min, and cooled to allow annealing at 53°C. A mixture containing Superscript III Reverse Transcriptase was added, and the reaction was continued at 53°C for an additional 20 min. Reactions were terminated by the addition of 2 × TBE-Urea sample buffer after incubation at 70°C for 15 min. Extension products were resolved on a 12.5% TBE-urea gel and visualized using a TyphoonFLA9000 photoimager (GE Healthcare).
Definition of A-site peaks
We first determined the A-site corresponding to each read by an offset of [(15/27)×(L)] from the 5' end of the read, where L is the length of each read. Then we used normalized A-site count (ribo/mRNA) as A-site peaks.
Definition of strong stalling
Strong stalling is defined as A-site peaks with a height of higher than eight because the number of A-site peaks in the ermBL region was around eight. This region is where ribosome stalling was confirmed by RNA foot printing. The results, however, remain robust also for other cutoff values of A-site density.
Enrichment of over-, under-represented peptide sequence
Over- and under-represented peptide sequences were identified as previously described [13], except that p-value cutoff is 0.0001 for under-represented peptides and 0.9999 for over-represented peptides.
Supplementary Information
Additional file 1: Supplementary Tables S1
Additional file 2: Supplementary Tables S2
Additional file 3: Supplementary Figure S1
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data generated in this study have been deposited to the DDBJ depository (DRA011224).
Competing interests
The authors declare that they have no competing interests.
Funding
This work was funded by Grant-in-Aid for Japan Society for the Promotion of Science Research Fellow 16J02984
Author contributions
ST contributed to the conception and overall design of the work, all of experiments, bioinformatics analysis and interpretation of Ribo-Seq data and drafting of the manuscript. TA contributed to the design of Ribo-Seq experiments. KY contributed to Ribo-Seq experiments. TH contributed to bioinformatics analysis, interpretation of Ribo-Seq data, drafting of the manuscript and substantial revision of the manuscript. YT contributed to the drafting of the manuscript. KH contributed to the conception and overall design of the work, interpretation of data, and drafting and revision the manuscript. The authors read and approved the final manuscript.
Acknowledgment
We thank Tomoko Yamamoto for discussions and comments to the manuscript.
References
- Xiong L, Shah S, Mauvais P, Mankin AS (1999) A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol 31: 633-639.
- Kannan K, Mankin AS (2011) Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann N Y Acad Sci 1241: 33-47.
- Sloan KE, Warda AS, Sharma S, Entian K-D, Lafontaine DLJ, et al. (2011) Modifications of ribosomal RNA: From enzymes to function. RNA Biol 14: 97-110.
- Baldridge KC, Contreras LM (2014) Functional implications of ribosomal RNA methylation in response to environmental stress. Crit Rev Biochem Mol Biol 49: 69-89.
- Nakatogawa H, Ito K (2002) The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629-636.
- Tenson T, Ehrenberg M (2002) Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591-594.
- Takaya A, Sato Y, Shoji T, Yamamoto T (2013) Methylation of 23S rRNA nucleotide G748 by RlmAII methyltransferase renders Streptococcus pneumoniae telithromycin susceptible. Antimicrob Agents Chemother 57: 3789-3796.
- Shoji T, Takaya A, Sato Y, Kimura S, Suzuki T, et al. (2015) RlmCD-mediated U747 methylation promotes efficient G748 methylation by methyltransferase RlmAII in 23S rRNA in Streptococcus pneumoniae; interplay between two rRNA methylations responsible for telithromycin susceptibility. Nucleic acids research 43: 8964-8972.
- Min Y-H, Kwon A-R, Yoon E-J, Shim MJ, Choi EC (2008) Translational attenuation and mRNA stabilization as mechanisms of erm (B) induction by erythromycin. Antimicrobial agents and chemotherapy 52: 1782-1789.
- Davis AR, Gohara DW, Yap MNF (2014) Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA 111: 15379-15384.
- Okada N, Tatsuno I, Hanski E, Caparon M, Sasakawa C (1998) Streptococcus pyogenes protein F promotes invasion of HeLa cells. Microbiology 144: 3079-3086.
- Takaya A, Kitagawa N, Kuroe Y, Endo K, Okazaki M, et al. (2010) Mutational analysis of reduced telithromycin susceptibility of Streptococcus pneumoniae isolated clinically in Japan. FEMS microbiology letters 307: 87-93.
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. (2007) "A" silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525-528.
- Tsai C-J, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, et al. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383: 281-291.
- Sabi R, Tuller T (2017) Computational analysis of nascent peptides that induce ribosome stalling and their proteomic distribution in Saccharomyces cerevisiae. Rna 23: 983-994.
- Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O'Brien EP (2013) In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci USA 110.: 132-140.
- Döring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, et al. (2017) Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170: 298-311.
- Richter JD, Coller J (2015) Pausing on polyribosomes: make way for elongation in translational control. Cell 163: 292-300.
- Moore SD, Sauer RT (2005) Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol Microbiol 58: 456-466.
- Charneski CA, Hurst LD (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoSBiol 11: 1001508.
- Liu TY, Song YS (2016) Prediction of ribosome footprint profile shapes from transcript sequences. Bioinformatics 32: 183-191.
- Wang M, Parshin AV, Shcherbik N, Pestov DG (2015) Reduced expression of the mouse ribosomal protein Rpl17 alters the diversity of mature ribosomes by enhancing production of shortened 5.8 S rRNA. RNA 21: 1240-1248.
- Zhang S, Hu H, Zhou J, He X, Jiang T (2017) Analysis of ribosome stalling and translation elongation dynamics by deep learning. Cell systems 5: 212-220.
- Kataja J, Seppälä H, Huovinen P (2001) In vitro activities of the novel ketolide telithromycin (hmr 3647) against erythromycin-resistant streptococcusspecies. Antimicrob Agents Chemother 45: 789-793.
- Nakhoul H, Ke J, Zhou X, Liao W, Zeng SX, et al. (2014) Ribosomopathies: mechanisms of disease. Clin Med Insights Blood Disord 7: 7-16.
- Narla A, Ebert BL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196-3205.
- Keersmaecker KD, Sulima SO, Dinman JD (2015) Ribosomopathies and the paradox of cellular hypo-to hyperproliferation. Blood 125: 1377-1382.
- Connolly M, Paul R, Farre-Garros R, Natanek SA, Bloch S, et al. (2018) miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J Cachexia Sarcopenia Muscle 9: 400-416.
- Hori H, Nakamura S, Yoshida F, Teraishi T, Sasayama D, et al. (2018) Integrated profiling of phenotype and blood transcriptome for stress vulnerability and depression. J Psychiatr Res 104: 202-210.
- Iannelli F, Pozzi G (2004) Method for introducing specific and unmarked mutations into the chromosome of Streptococcus pneumoniae. Molecular biotechnology 26: 81-86.
- Bolger A, Giorgi F (2014) Trimmomatic: a flexible read trimming tool for illumina NGS data. Bioinformatics 30: 2114-2120.
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37: 907-915.
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 17: 10-12.
- Basu A, Yap MNF (2016) Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic acids research 44: 4881-4893.
- Sigmund CD, Ettayebi M, Borden A, Morgan EA (1988) Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods in enzymology. Academic Press 164: 673-690.
Supplementary Files
|
Stalling peptides in Sp284 |
Stalling peptides in Sp379 |
Common Stalling peptides |
|
AAA |
AAR |
EYN |
|
AAC |
AAV |
KVQ |
|
AAD |
ADQ |
PKP |
|
AAE |
AEE |
ERG |
|
AAI |
AEK |
EKP |
|
AAK |
AFN |
KKV |
|
AAL |
AKA |
NGR |
|
AAP |
AKK |
GDF |
|
AAR |
APA |
KAA |
|
AAT |
APK |
QQP |
|
AAV |
APP |
NSA |
|
ADN |
APQ |
NWG |
|
ADQ |
ARQ |
DFA |
|
AEE |
ARR |
TGK |
|
AEI |
ATD |
KKA |
|
AEK |
ATH |
LGR |
|
AEQ |
ATR |
VAV |
|
AFR |
AVA |
TPA |
|
AGA |
CCN |
EYC |
|
AGP |
CNR |
WGD |
|
AHA |
CNW |
AEE |
|
AHI |
DFA |
EPE |
|
AHV |
DGP |
AAV |
|
AIK |
DHG |
RRV |
|
AKA |
DHN |
APK |
|
AKG |
DIP |
YTG |
|
AKK |
DQM |
TAA |
|
AKP |
DQQ |
TDG |
|
AKR |
DTG |
ERK |
|
AKV |
EAD |
PHT |
|
ANE |
EAV |
KPA |
|
ANP |
EEY |
RVG |
|
APA |
EHK |
TQQ |
|
APE |
EKP |
WYV |
|
APG |
EKS |
VDH |
|
APK |
EKT |
IRH |
|
APQ |
EPE |
PVR |
|
APV |
ERG |
GAA |
|
AQW |
ERK |
HKK |
|
ARG |
EYC |
HEG |
|
ARQ |
EYN |
RRS |
|
ARR |
FAK |
PKA |
|
AST |
FTG |
KRH |
|
ATD |
FTK |
ARQ |
|
ATE |
FVN |
AVA |
|
ATG |
GAA |
ADQ |
|
ATH |
GCN |
IPS |
|
ATR |
GDF |
DQQ |
|
ATT |
GGC |
GCN |
|
AVA |
GHI |
DHG |
|
AVE |
GKR |
RRL |
|
AVG |
GKV |
SLT |
|
AVK |
GPH |
EAV |
|
AVN |
GQV |
KRA |
|
AVS |
GRR |
PEK |
|
AYT |
GSY |
QQA |
|
CNW |
GTG |
PAP |
|
CTT |
HEG |
RGG |
|
DAA |
HGH |
GGC |
|
DFA |
HGK |
KAE |
|
DGP |
HIL |
RRA |
|
DHG |
HKE |
KVW |
|
DNE |
HKK |
AEK |
|
DPH |
HRR |
QPP |
|
DQQ |
HSF |
MKR |
|
DRW |
HVD |
RHR |
|
DTA |
IEP |
CNW |
|
DTG |
IGH |
AAR |
|
DVE |
IKT |
PTP |
|
DWM |
INV |
MAK |
|
DYA |
IPS |
AKK |
|
EAD |
IRH |
HVD |
|
EAN |
KAA |
INV |
|
EAP |
KAE |
DGP |
|
EAV |
KCC |
SAN |
|
EET |
KDV |
RKH |
|
EGA |
KKA |
PAE |
|
EGD |
KKV |
APQ |
|
EGG |
KNK |
RGK |
|
EGP |
KPA |
GTG |
|
EGV |
KPK |
GSY |
|
EHR |
KRA |
PPK |
|
EIN |
KRG |
ATR |
|
EKA |
KRH |
TGS |
|
EKP |
KRV |
RGL |
|
ENN |
KTE |
QQQ |
|
EPE |
KVQ |
APA |
|
EQF |
KVW |
ARR |
|
EQH |
KWY |
DTG |
|
EQP |
LDG |
NEP |
|
ERG |
LGR |
SAQ |
|
ERK |
MAK |
EAD |
|
ETH |
MKR |
YAR |
|
ETP |
MRV |
ATD |
|
ETR |
MVE |
AKA |
|
EVA |
NAV |
ATH |
|
EVN |
NDA |
HRR |
|
EYC |
NEP |
RVT |
|
EYN |
NGR |
|
|
FRA |
NHG |
|
|
FRR |
NRH |
|
|
GAA |
NSA |
|
|
GAK |
NVK |
|
|
GCN |
NVV |
|
|
GDF |
NWG |
|
|
GEG |
PAE |
|
|
GGA |
PAP |
|
|
GGC |
PEK |
|
|
GGG |
PGQ |
|
|
GGR |
PHR |
|
|
GGS |
PHT |
|
|
GHA |
PKA |
|
|
GIK |
PKK |
|
|
GIT |
PKP |
|
|
GKA |
PPK |
|
|
GPK |
PPV |
|
|
GPT |
PSY |
|
|
GRK |
PTP |
|
|
GRY |
PVR |
|
|
GSA |
QPP |
|
|
GSV |
QPS |
|
|
GSY |
QQA |
|
|
GTE |
QQP |
|
|
GTG |
QQQ |
|
|
GTW |
REK |
|
|
GVA |
RGG |
|
|
GVK |
RGK |
|
|
GYN |
RGL |
|
|
HAA |
RHR |
|
|
HAD |
RKH |
|
|
HEG |
RNT |
|
|
HID |
RQA |
|
|
HIW |
RQF |
|
|
HKK |
RRA |
|
|
HQR |
RRL |
|
|
HRR |
RRQ |
|
|
HRT |
RRS |
|
|
HTD |
RRV |
|
|
HVD |
RSE |
|
|
HVE |
RVD |
|
|
HVP |
RVG |
|
|
HWI |
RVR |
|
|
IAA |
RVT |
|
|
IIA |
RYT |
|
|
INV |
SAN |
|
|
IPS |
SAQ |
|
|
IRH |
SKN |
|
|
ITI |
SLT |
|
|
IVE |
SPQ |
|
|
KAA |
SYR |
|
|
KAE |
TAA |
|
|
KAG |
TDG |
|
|
KAH |
TEG |
|
|
KAV |
TEH |
|
|
KAY |
TGK |
|
|
KEV |
TGS |
|
|
KGG |
TKN |
|
|
KGS |
TPA |
|
|
KGT |
TPG |
|
|
KKA |
TPT |
|
|
KKG |
TQQ |
|
|
KKP |
TRR |
|
|
KKV |
TSK |
|
|
KPA |
TWE |
|
|
KPG |
VAG |
|
|
KRA |
VAV |
|
|
KRF |
VCY |
|
|
KRH |
VDH |
|
|
KSA |
VDI |
|
|
KSK |
VEP |
|
|
KTK |
VKP |
|
|
KTV |
VNH |
|
|
KVQ |
VNI |
|
|
KVW |
VTP |
|
|
KYE |
VWI |
|
|
LAR |
WAT |
|
|
LGR |
WEI |
|
|
LKA |
WGD |
|
|
LRR |
WIK |
|
|
LTA |
WYV |
|
|
MAH |
YAR |
|
|
MAK |
YCT |
|
|
MAN |
YNR |
|
|
MDN |
YRG |
|
|
MDW |
YRV |
|
|
MKR |
YTG |
|
|
MLA |
|
|
|
MNT |
|
|
|
MQP |
|
|
|
MRG |
|
|
|
MRT |
|
|
|
NAA |
|
|
|
NAE |
|
|
|
NAK |
|
|
|
NEM |
|
|
|
NEP |
|
|
|
NES |
|
|
|
NGA |
|
|
|
NGR |
|
|
|
NKP |
|
|
|
NNH |
|
|
|
NQP |
|
|
|
NSA |
|
|
|
NTK |
|
|
|
NTT |
|
|
|
NVN |
|
|
|
NVP |
|
|
|
NWG |
|
|
|
PAE |
|
|
|
PAL |
|
|
|
PAN |
|
|
|
PAP |
|
|
|
PEA |
|
|
|
PEK |
|
|
|
PEP |
|
|
|
PGG |
|
|
|
PGH |
|
|
|
PGS |
|
|
|
PHE |
|
|
|
PHT |
|
|
|
PHV |
|
|
|
PKA |
|
|
|
PKP |
|
|
|
PKV |
|
|
|
PMA |
|
|
|
PPH |
|
|
|
PPK |
|
|
|
PQP |
|
|
|
PSW |
|
|
|
PTP |
|
|
|
PTR |
|
|
|
PTS |
|
|
|
PVA |
|
|
|
PVR |
|
|
|
PWR |
|
|
|
QAE |
|
|
|
QMD |
|
|
|
QNE |
|
|
|
QPE |
|
|
|
QPK |
|
|
|
QPP |
|
|
|
QPT |
|
|
|
QQA |
|
|
|
QQP |
|
|
|
QQQ |
|
|
|
QRA |
|
|
|
QRV |
|
|
|
QSG |
|
|
|
QVA |
|
|
|
QWR |
|
|
|
RAK |
|
|
|
RAR |
|
|
|
RAS |
|
|
|
RDS |
|
|
|
REF |
|
|
|
RFR |
|
|
|
RGG |
|
|
|
RGI |
|
|
|
RGK |
|
|
|
RGL |
|
|
|
RHG |
|
|
|
RHR |
|
|
|
RHT |
|
|
|
RHY |
|
|
|
RIN |
|
|
|
RKH |
|
|
|
RPH |
|
|
|
RPR |
|
|
|
RPT |
|
|
|
RQG |
|
|
|
RQK |
|
|
|
RRA |
|
|
|
RRE |
|
|
|
RRL |
|
|
|
RRN |
|
|
|
RRP |
|
|
|
RRS |
|
|
|
RRV |
|
|
|
RSP |
|
|
|
RTA |
|
|
|
RTG |
|
|
|
RVE |
|
|
|
RVG |
|
|
|
RVT |
|
|
|
RWN |
|
|
|
SAA |
|
|
|
SAN |
|
|
|
SAQ |
|
|
|
SAV |
|
|
|
SCT |
|
|
|
SHR |
|
|
|
SLT |
|
|
|
STK |
|
|
|
SVA |
|
|
|
SVM |
|
|
|
TAA |
|
|
|
TAH |
|
|
|
TAQ |
|
|
|
TDG |
|
|
|
TDT |
|
|
|
TDV |
|
|
|
TEA |
|
|
|
TGA |
|
|
|
TGK |
|
|
|
TGP |
|
|
|
TGR |
|
|
|
TGS |
|
|
|
TGT |
|
|
|
TGW |
|
|
|
THK |
|
|
|
TKA |
|
|
|
TKE |
|
|
|
TKT |
|
|
|
TPA |
|
|
|
TPH |
|
|
|
TPR |
|
|
|
TQQ |
|
|
|
TRL |
|
|
|
TSH |
|
|
|
TST |
|
|
|
TTT |
|
|
|
TVV |
|
|
|
TWQ |
|
|
|
TYK |
|
|
|
VAA |
|
|
|
VAE |
|
|
|
VAS |
|
|
|
VAV |
|
|
|
VDH |
|
|
|
VEA |
|
|
|
VEG |
|
|
|
VEK |
|
|
|
VES |
|
|
|
VEY |
|
|
|
VGE |
|
|
|
VGG |
|
|
|
VGR |
|
|
|
VGT |
|
|
|
VIP |
|
|
|
VIS |
|
|
|
VKA |
|
|
|
VKK |
|
|
|
VKT |
|
|
|
VKV |
|
|
|
VMD |
|
|
|
VNV |
|
|
|
VVF |
|
|
|
WGD |
|
|
|
WIT |
|
|
|
WKQ |
|
|
|
WME |
|
|
|
WRG |
|
|
|
WRQ |
|
|
|
WYV |
|
|
|
WYY |
|
|
|
YAH |
|
|
|
YAR |
|
|
|
YCK |
|
|
|
YED |
|
|
|
YEH |
|
|
|
YET |
|
|
|
YNA |
|
|
|
YQP |
|
|
|
YTG |
|
|
|
YVV |
|
|
Table S1: The list of stalling peptides.
The first and the second columns list stalling peptides in Sp284 and Sp379 respectively. The third column shows the list of stalling peptides that are common between strains.
|
Over-represented prptides |
Under-represented peptides |
|
ACG |
AAE |
|
ACS |
AAP |
|
ADF |
AAT |
|
ADK |
ADP |
|
ADY |
ADT |
|
AEK |
AEP |
|
AGF |
AES |
|
AGI |
AGP |
|
AGL |
AKF |
|
AGV |
ANE |
|
AIL |
APP |
|
AKE |
ASE |
|
AKN |
AVP |
|
ALA |
CCC |
|
AQA |
CCD |
|
ASH |
CCW |
|
ASQ |
CHC |
|
ATG |
CHW |
|
AYQ |
CKC |
|
CFE |
CMQ |
|
CHP |
CMR |
|
CHY |
CMW |
|
CSK |
CPM |
|
DEV |
CQC |
|
DFD |
CVW |
|
DFE |
CWP |
|
DFF |
CYW |
|
DFI |
DAN |
|
DFL |
DAQ |
|
DFP |
DDE |
|
DFS |
DDG |
|
DFV |
DDN |
|
DFY |
DDP |
|
DGK |
DDR |
|
DGQ |
DDT |
|
DGT |
DEG |
|
DIP |
DGG |
|
DKI |
DGP |
|
DKV |
DHD |
|
DLA |
DHK |
|
DLD |
DIG |
|
DLP |
DND |
|
DNL |
DNE |
|
DNP |
DNK |
|
DQV |
DPG |
|
DRI |
DPP |
|
DYF |
DRE |
|
DYH |
DRG |
|
DYI |
DSA |
|
DYL |
DSE |
|
DYQ |
DTE |
|
DYY |
DTG |
|
EAG |
DTK |
|
EAI |
DTQ |
|
EDF |
DVG |
|
EDL |
DVS |
|
EEA |
EDD |
|
EEI |
EDE |
|
EEL |
EDN |
|
EEV |
EDP |
|
EIA |
EDQ |
|
EIL |
EDR |
|
EKE |
EDS |
|
EKG |
EDT |
|
EKI |
EEP |
|
EKL |
EFK |
|
EKM |
EGE |
|
EKR |
EGG |
|
EKT |
EGP |
|
EKV |
EGS |
|
EKY |
EHE |
|
ELA |
EPA |
|
ENG |
EPD |
|
ENI |
EPE |
|
ENL |
EPG |
|
ENP |
EPK |
|
ENQ |
EPN |
|
EQA |
EPP |
|
EQI |
EPQ |
|
EQV |
EPR |
|
ERF |
EQH |
|
ERI |
EQN |
|
ERL |
EQQ |
|
ETG |
EQR |
|
EVV |
ESA |
|
FAK |
ESE |
|
FCQ |
ESK |
|
FDE |
ESN |
|
FDK |
EST |
|
FDN |
ETQ |
|
FDQ |
EWD |
|
FEE |
EYE |
|
FEN |
EYK |
|
FER |
EYN |
|
FKN |
EYT |
|
FNP |
FEY |
|
FNQ |
FII |
|
FSD |
FIM |
|
FSP |
FKF |
|
FVT |
FKI |
|
FWN |
FKL |
|
FYQ |
FKV |
|
GAG |
FKY |
|
GCG |
FLI |
|
GCS |
FMF |
|
GFD |
FML |
|
GFS |
FMS |
|
GIP |
FMY |
|
GIS |
FQL |
|
GKI |
FRL |
|
GKS |
FRV |
|
GKT |
FYV |
|
GLP |
GAA |
|
GLT |
GAE |
|
GNP |
GAQ |
|
GQT |
GCW |
|
GQV |
GDA |
|
GRI |
GDD |
|
GSG |
GDP |
|
GTG |
GEA |
|
GTP |
GEG |
|
GVD |
GEQ |
|
HCF |
GGD |
|
HFS |
GKA |
|
HFT |
GLQ |
|
HHF |
GNE |
|
HHL |
GNK |
|
HPD |
GPA |
|
HPE |
GPD |
|
HYH |
GPE |
|
HYP |
GPF |
|
HYQ |
GPI |
|
IAG |
GPK |
|
IAQ |
GPL |
|
IAR |
GPP |
|
IAS |
GPQ |
|
IEE |
GTN |
|
IEK |
GTQ |
|
IEN |
GWC |
|
IFP |
HCM |
|
IIG |
HCW |
|
IKE |
HDD |
|
ILA |
HED |
|
ILP |
HND |
|
ILS |
HNE |
|
INQ |
HNN |
|
IPA |
HWW |
|
IPE |
IEF |
|
IPN |
IEI |
|
IPT |
IEP |
|
IPV |
IEY |
|
IQE |
III |
|
IRD |
IIK |
|
IRK |
IIL |
|
IRQ |
IIY |
|
ISQ |
IKF |
|
ISR |
IKI |
|
ISS |
IKL |
|
IYQ |
IKM |
|
KDF |
IKV |
|
KDG |
IKY |
|
KDL |
ILI |
|
KDY |
ILY |
|
KEA |
IMF |
|
KEE |
IMI |
|
KEI |
IML |
|
KEK |
IMY |
|
KEL |
INI |
|
KEN |
IQI |
|
KEV |
ITL |
|
KHC |
IVV |
|
KIA |
IWC |
|
KKE |
IYI |
|
KKI |
KCE |
|
KKV |
KCK |
|
KKW |
KDD |
|
KNG |
KFE |
|
KNP |
KFF |
|
KNR |
KFK |
|
KQV |
KFL |
|
KRI |
KFM |
|
KRL |
KFP |
|
KRV |
KFQ |
|
KTI |
KFR |
|
KTV |
KFS |
|
KWY |
KFT |
|
LAD |
KGA |
|
LAE |
KGP |
|
LAG |
KGS |
|
LAK |
KHE |
|
LAQ |
KHK |
|
LAS |
KHT |
|
LDE |
KLM |
|
LDK |
KLQ |
|
LDY |
KME |
|
LEE |
KPP |
|
LEK |
KQQ |
|
LEN |
KSA |
|
LGI |
KSE |
|
LKD |
KYT |
|
LKE |
KYV |
|
LKK |
LEF |
|
LKN |
LEL |
|
LLA |
LEP |
|
LNP |
LEY |
|
LNQ |
LFI |
|
LPE |
LFK |
|
LPF |
LFM |
|
LPS |
LGE |
|
LPV |
LGP |
|
LPY |
LHV |
|
LRE |
LHW |
|
LSD |
LII |
|
LSG |
LIK |
|
LSK |
LIL |
|
LSN |
LIM |
|
LSQ |
LKC |
|
LSR |
LKF |
|
LTD |
LKI |
|
LTE |
LKL |
|
LTP |
LKM |
|
LYQ |
LKV |
|
MKE |
LKY |
|
MKI |
LLI |
|
MKK |
LLL |
|
MKQ |
LLM |
|
MKR |
LLW |
|
MLE |
LLY |
|
MNQ |
LMF |
|
MRQ |
LML |
|
MSK |
LMY |
|
MSR |
LNF |
|
MTK |
LQF |
|
MTS |
LQI |
|
NGI |
LQL |
|
NGK |
LQM |
|
NGQ |
LQQ |
|
NGR |
LQW |
|
NHQ |
LRI |
|
NIP |
LRL |
|
NLP |
LRM |
|
NLS |
LRT |
|
NLT |
LRV |
|
NPA |
LVI |
|
NPD |
LWV |
|
NPE |
LYI |
|
NPF |
LYM |
|
NPK |
LYV |
|
NPN |
MCH |
|
NPQ |
MCW |
|
NPS |
MEP |
|
NPT |
MFI |
|
NPY |
MHV |
|
NQE |
MLI |
|
NQF |
MLL |
|
NQL |
MLY |
|
NQV |
MYC |
|
NRF |
MYE |
|
NRL |
MYF |
|
NRV |
MYI |
|
NRY |
MYK |
|
NYL |
MYV |
|
|
NAT |
|
PEE |
NDD |
|
PEK |
NDS |
|
PEN |
NDT |
|
PIL |
NED |
|
PKT |
NEF |
|
PNC |
NEG |
|
PSQ |
NEH |
|
PVF |
NEK |
|
PVI |
NES |
|
PVL |
NGP |
|
PVY |
NIN |
|
QAG |
NKF |
|
QAI |
NND |
|
QAL |
NNE |
|
QAY |
NSA |
|
QDL |
NSM |
|
QEI |
NST |
|
QEK |
PCY |
|
QEL |
PFR |
|
QGI |
PGE |
|
QKI |
PGL |
|
QTI |
PKF |
|
QTV |
PLK |
|
QVA |
PLQ |
|
QVL |
PLR |
|
RDY |
PPA |
|
RFG |
PPD |
|
RQA |
PPE |
|
RYF |
PPG |
|
RYQ |
PPK |
|
SGG |
PPP |
|
SGK |
PPQ |
|
SKE |
PPV |
|
SKH |
PQR |
|
SLA |
PYE |
|
SLS |
QCC |
|
SPE |
QGP |
|
SQA |
QKS |
|
SQE |
QND |
|
SQT |
QPG |
|
SSS |
QQD |
|
SWY |
QQN |
|
SYL |
QQS |
|
TDE |
QSE |
|
TGK |
QSN |
|
TGW |
QWC |
|
THF |
QYK |
|
TIA |
RAD |
|
TIL |
RAP |
|
TPD |
RCC |
|
TPE |
REP |
|
TPV |
RGD |
|
TSP |
RGP |
|
TVE |
SAA |
|
TVL |
SAG |
|
TWY |
SAN |
|
VAT |
SDA |
|
VDG |
SED |
|
VEK |
SEL |
|
VEN |
SEP |
|
VKD |
SES |
|
VKE |
SGP |
|
VLA |
SNE |
|
VLD |
SPG |
|
VLP |
SPP |
|
VLS |
STE |
|
VNP |
TEG |
|
VPV |
TEY |
|
VSS |
TGP |
|
VTS |
TKF |
|
VVD |
TKG |
|
WEN |
TKL |
|
WKE |
TKM |
|
WQE |
TKS |
|
WYQ |
TLQ |
|
WYY |
TMY |
|
YCH |
TQL |
|
YDF |
TRS |
|
YFD |
TTQ |
|
YHF |
VEY |
|
YHP |
VII |
|
YLD |
VIL |
|
YPD |
VKF |
|
YQA |
VKI |
|
YQD |
VKL |
|
YQE |
VKM |
|
YQG |
VLI |
|
YQH |
VLL |
|
YQK |
VQL |
|
YQL |
VVL |
|
YQN |
VYK |
|
YQT |
WAA |
|
YQV |
WCC |
|
YRE |
WCE |
|
YRK |
WCH |
|
YYL |
WCT |
|
YYQ |
WHC |
|
|
WPC |
|
|
WPG |
|
|
WPH |
|
|
WPW |
|
|
WQC |
|
|
WWC |
|
|
WWM |
|
|
WWP |
|
|
WWW |
|
|
YCC |
|
|
YEF |
|
|
YEL |
|
|
YEY |
|
|
YIV |
|
|
YKI |
|
|
YKL |
|
|
YKS |
|
|
YKY |
|
|
YML |
|
|
YVI |
|
|
YVM |
|
|
YWC |
Table S2: The list of over- and under-represented peptides.
The first and the second columns list over-represented peptides and under-represented peptides in S. pneumoniae proteome composition.
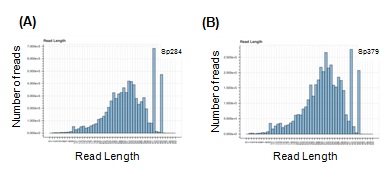 Figure S1: Histograms showing the distribution of length of RFPs in Sp284 and Sp379, respectively.
Figure S1: Histograms showing the distribution of length of RFPs in Sp284 and Sp379, respectively.
Citation: Shoji T, Takaya A, Kusuya Y, Takahashi H, Kawashima H (2021) Ribosome Profiling in Streptococcus pneumoniae reveals the Role of Methylation of 23S rRNA Nucleotide G748 on Ribosome Stalling. J Genet Genomic Sci 6: 024.
Copyright: © 2021 Tatsuma Shoji, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

